Research about Alternative Splicing recently 楊佳熒.
-
Upload
pamela-adams -
Category
Documents
-
view
231 -
download
0
Transcript of Research about Alternative Splicing recently 楊佳熒.

Research about Alternative Splicing recently
楊佳熒

Transcription Start
ATG (Met) TAATAGTGA
Translation Start Stop codon
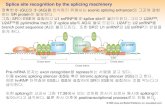
Intron 1 Intron 25‘ 3‘Exon 1 Exon 2 Exon 3
Acceptor splice site
Donor splice site
Intron 1 Intron 2Exon 1 Exon 2 Exon 3AGGT AGGT
mRNA Exon 1 Exon 2 Exon 33‘UTR
5‘UTR
ORF
Protein
Features of a Mammalian Gene
Gene start Gene end

Introduce NCBI (Nation Center for Biotechnology Information) refseq ----Genome Annotation

Known Gene20660
Predicted Gene6912

<ex>

Alternative Splicing

Research about alternative splicing

NCBI(National Center of Biotechnology Information)
TIGR(The Institute of Genomic Research)
UniGene
HGI(Human Gene Index)
alignment
Genome sequences

cDNA
cDNA (complement DNA)
EST (Expressed Sequence Tag) DB
TIGR(The Institute of Genomic Research)
NCBI(National Center of Biotechnology Information)
Data Source
HGI(Human Gene Index) UniGene



An alternative splicing example

Classification of alternative splicing

Two question
1. How many are the result of aberrant splicing or noise data ?
2. How many of these predicted splice variants are functional ?

A Comparative Method for Identification of Gene Structures and Alternatively Spliced Variants
Trees-Juen Chuang, Feng-Chi Chen and Meng-Yuan Chou
Bioinformatics Advance Access published June 24, 2004
Question 1: How many are the result of aberrant splicing or noise data ?Method 1 : 修補



Genome wide identification and classification of alternative splicing based on EST data
S.Gupta, D.Zink, B.Korn, M.Vingron, S.A.Haas
Bioinformatics Advance Access published April 29, 2004
Question 1: How many are the result of aberrant splicing or noise data ?Method 2 : 設定很嚴格的條件限制

Flow chart
UniGene 1 assembly 2 GeneNest
3 Mapping to Genome(sim4) 4 SpliceNest

Four parameters
1. Alignment Quality Alignment quality is computed based on the sim4.
2. Signal Information (splice signal) (1) the presence of the consensus (GT-AG) splice
signal
(2) the presence of a non-consensus splice signal with perfect alignment in vicinity (+/-10 bases)

3. Common Boundaries Common boundaries observed among related exons of
different transcripts provides further evidence for a splice junction.
4. Neighborhood Parameters overall splicing direction


How prevalent is functional alternative splicing in the human genome ?
Rotem Sorek, Ron Shamir and Gil Ast
TRENDS in Genetics Vol.20 No.2 February 2004
Question 2: How many of these predicted splice variants are functional ?

Method
Finding exon-skipping (cassette exons) that are conserved between humans and mouse
1. Identification of mouse ESTs that contain the exon and the two flanking exons.
2. If the exon was not represented in mouse ESTs, the sequence of the human exon was searched against the intron by the skipping mouseEST on the mouse genome.


Conclude
1. We conclude that a signification portion of cassette exons evidence in EST database is not functional.

Idea
1. Discovery of novel splice forms and functional analysis of cancer-s
pecific alternative splicing in human expressed sequences
Qiang Xu and Christopher LeeNucleic Acids Research, 2003, Vol.31 No.19

EGFR gene
DWER


2.
EST analysis online: WWW tools for detection of SNPs and alternative splice forms
David Brett, Gerrit Lehmann, Jens Hanke, Stefan Gross,
Jens Reich and Peer Bork
Trends in Genetics, Volume 16, Issue 9, 1 September 2000,