Step
description
Transcript of Step

’5
Step 1
Step 4
BHQ1
TTG
TTG
F
BHQ1
AAC
5’3’
5’3’
F
BHQ1
TTG3’
Step 1
Step 4
BHQ1
TTG
TTG
F
BHQ1
5’3’
5’3’
BHQ1
TTG3’
BHQ1
TTG3’
gene mutated : gene wild type :
GGT5’ 3’CCAPNA:
amplification NO amplification
mutated : wild type :
5’ 3’
Step 1
Step 4
BHQ1
TTG
TTG
F
BHQ1
5’3’
5’3’
BHQ1
TTG3’
BHQ1
TTG3’
Step 1
Step 4
BHQ1
TTG
F
BHQ1
5’3’
5’3’
BHQ1
TTG3’
BHQ1
TTG3’
BHQ1
TTG3’
mutated : wild type :
5’ 3’
amplification NO amplification
mutated : wild type :
5’ 3’GTT5’ 3’GTT5’ 3’GTT5’ 3’GTT5’ 3’
Step 2
Step 3
RE
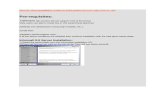
Asymmetric PNA-PCR/OCEAN for Kras mutation detection
AAC
FFFF
AAC
Figure S1 . PNA-PCR/OCEAN method for K-ras mutation detection.
Assymetric PNA-PCR: a PNA probe complementary to the wild type gene suppresses amplification of the codon 12 wild type K-ras sequence and amplifies preferentially mutant sequences. The PCR reaction is performed using an imbalanced forward:reverse primer ratio in order to preferentially amplify the strand which is complementary to the OCEAN probes. OCEAN: steps 1-4 are carried out in a single tube. Step 1: a stabilizing probe (anchor, in dark gray) binds to the amplified strand of K-ras; step 2: a second probe (amplifier, in light gray, carrying a Fluorescein and a Black Hole Quencher, BHQ1) complementary to the mutant gene recognizes and binds the duplex forming a three-way junction. The resulting ternary structure contains the recognition site for a restriction endonuclease (RE). Step 3: The endonuclease cleaves the amplifier. The cleavage separates the fluorophore from the Black Hole Quencher with consequent emission of fluorescence. Step 4: the cleaved amplifier dissociates, and the cycle repeats itself thus resulting in a continuous signal amplification. For each sample, 4 different OCEAN reactions are performed, each containing a different labelled amplifier, specific for the 4 most frequent mutations of K-ras codon 12.
S
S
S